This week I had the opportunity to extract DNA from e. coli under Paul's
supervision and guidance. Although his method is pretty much fool proof I was
super excited to see the banding on our gels! I honestly though my first time
would be a disaster hehe. The protocol followed is summarized below.
1. Centrifuge 1mL of cultured TSB @ 12000
rpm for 3 minutes.
2. Discard supernatant (broth) & add
0.3 mL TBS & 15 µL 10% SDS to each tube.
3. Incubate @ 55ºC for 10 min.
4. Cool to room temp
5. Add 5µL proteinase K -
Vortex 20 sec.
6. Incubate in ice for 5 min.
7. Centrifuge @12000 rmp for 3 min. (to precipitate proteins)
8. Transfer supernatant to clean 1.5 mL tubes.
9. Add 300µL room temp isopropanol.
10. Invert until DNA forms threads.
11. Centrifuge @ 12000 rpm – 2 min.
12. Discard supernatant. Add 300µL room temp 70% ethanol. Invert
to wash pellet.
13. Centrifuge @ 12000 rpm – 2 min.
14. Discard supernatant & air dry DNA (we used the hood for
20min.)
15. Resuspend DNA in sterile water (50µL).
While the samples were incubating for step 3 above, we prepared 1xTAE
buffer by adding 980 mL of di water to 20 mL of TAE 0.5%.
While DNA was drying under the hood, we prepared the gels for
electrophoresis by adding 0.9 g of agarose to 100mL of 1x TAE buffer. We
proceeded to microwave this beaker for 30 sec intervals until diluted. Once it
cooled down to 59ºC we added 4µL of cyber green.
We then poured 25mL of agarose/TAE buffer solution into the gel molds
we had set up previously and covered with box to keep in the dark since cyber
green is very light sensitive.
Once the gels were set and the DNA had dried, we completed step 15
of the above protocol and proceeded to load 4µL of DNA sample and 2µL of
loading dye in the wells in the gel.
The last step was to run electricity (120V) through the gels for
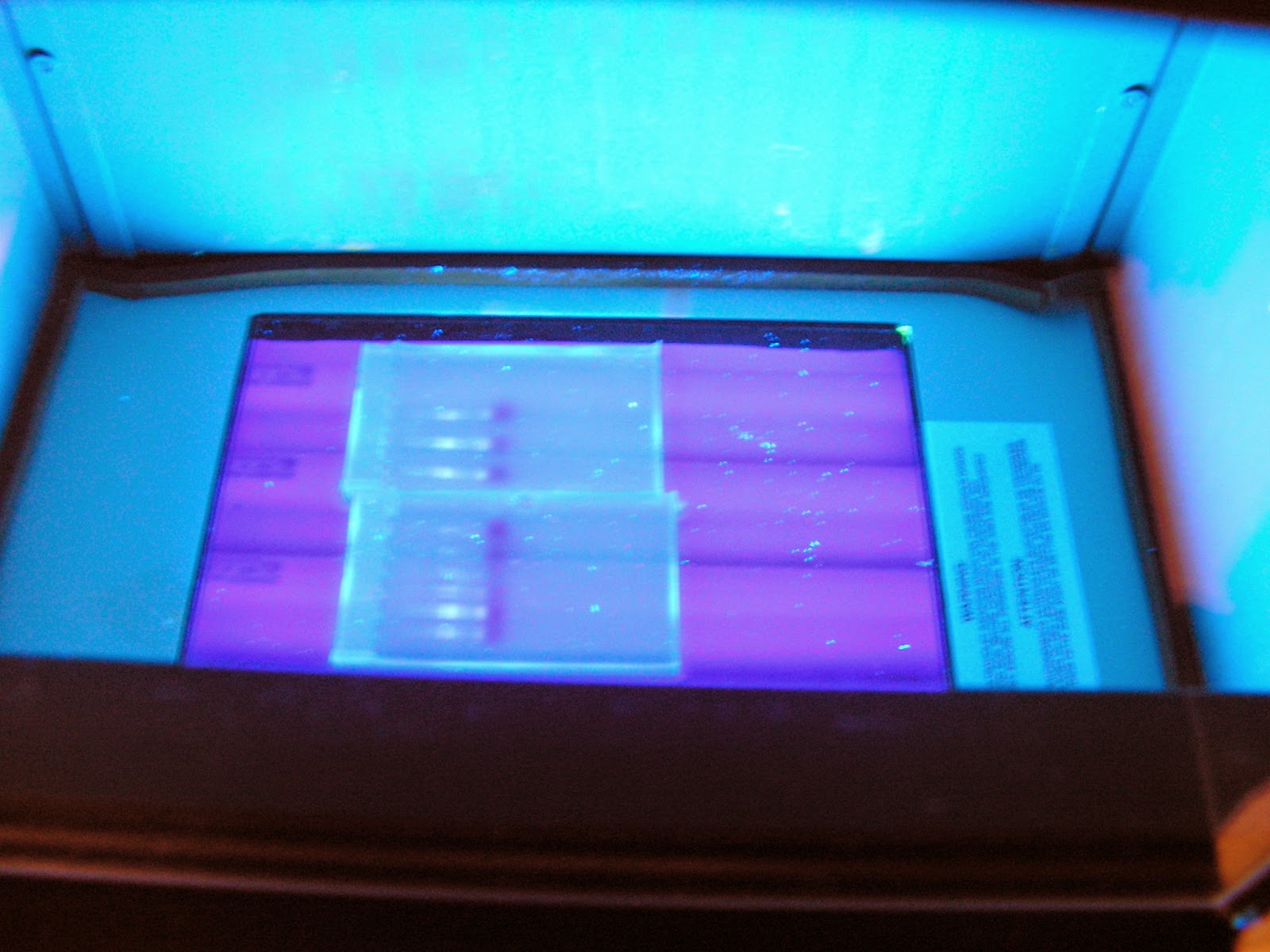
approximately 15 min. And view them under UV light. It was a success!! Please
see pictures below.
Thank you very much to Paul Cattelino for your help and all the
little tips you shared with me! I love being part of this program and feel
honored to being able to work alongside and collaborate with you!!
Muchas Gracias Paul!
DNA threads precipitated with isopropanol
Gels undergoing electrophoresis
Left to right: Sample 1, Sample 2 and control (Paul's older extraction)
Another view with second gel added to the right. From left to right on second gel:
Sample 1 and Sample 2. Very nice banding!!!









.jpeg)
.jpeg)
.jpeg)
.jpeg)


